Introduction




Introduction
Comparing behavior across individuals
Introduction
Goal
We want to evaluate how similar is the Spontaneous Brain Activity across individuals
- record Spontaneous Brain Activity → multiple individuals + whole-brain + neuron-scale
- find a shared representation of this activity across fish
- build of vocabulary of brain states
- compare the structure of the sequence of states
Introduction
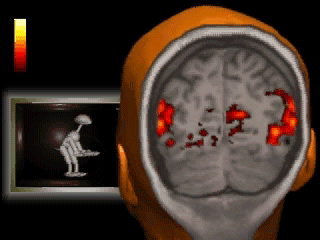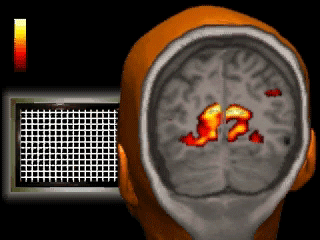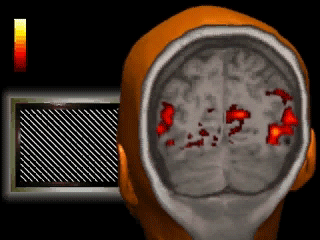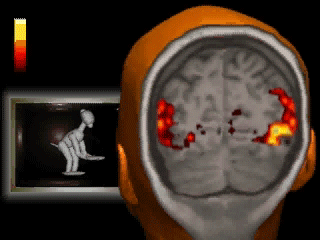
Introduction
Introduction
Introduction
Introduction
Introduction
Zebrafish


- Rich behavior
- Transparent
- Small brain (~100k neurons)
- Genetic tools (GCaMP, ...)
Introduction
Light Sheet Microscopy
Introduction
Light Sheet Microscopy
Restricted Boltzmann Machines
Theory : A Probabilistic Model
Geoffrey Hinton

visible configuration
$$\mathbf v=(v_1,\dots,v_N)\in\{\text{on},\text{off}\}^N$$hidden configuration
$$\mathbf h=(h_1,\dots,h_M)\in\mathbb{R}^M$$Restricted Boltzmann Machines
Theory : A Generative Model
Restricted Boltzmann Machines
from zebrafish larvae
Thijs van der Plas
Jérôme Tubiana
Restricted Boltzmann Machines
Zebrafish Brain as a Composition of Cell Assemblies
Thijs van der Plas
Jérôme Tubiana
cell assembly = group of co-activating neurons, usually spatially localized
Restricted Boltzmann Machines
Zebrafish Brain as a Composition of Cell Assemblies
Thijs van der Plas
Jérôme Tubiana

Restricted Boltzmann Machines
provide Degenerate Representations
Goal : Find a Common Representation for the Spontaneous Brain Activity of multiple fish.
- Hidden Units ≈ known Functional Networks
- Functional Networks are shared by all individuals
⠀
⇒ multiple RBMs shoud provide the same representation
RBMs are stochastic ⇒ variable solutions
Restricted Boltzmann Machines
Summary
Goal : Find a Common Representation for the Spontaneous Brain Activity of multiple fish.
Spontaneous Brain Activity ⇒ no alignment
RBMs representation ⇒ composition of cell assemblies
RBMs representation ⇒ degenerate
⇒ We needs a method which explicitely aligns the latent space across individuals
Aligning Latent Space across Individuals
Two Methods
Neuronal Models With Shared Hidden Space
At the single neuron scale ?
⠀
Problem : neurons are not comparable across individuals
Hypothesis : cell assemblies are shared across individuals
Multi-fish Neuronal Model
Latent-aligned RBMs : Method
Multi-fish Neuronal Model
Latent-aligned RBMs : Method
Jorge Fernández
⠀
de Cossío Díaz

- Converge faster (1/10th)
- Converge more reliably
Multi-fish Neuronal Model
Hidden Units map to Stereotypic Neuronal Populations
Multi-fish Neuronal Model
Hidden Units describe a Shared Space
Multi-fish Neuronal Model
Translating Activity from one Fish to Another
- Shared latent space across multiple individuals
- Generative Model
⇒ Translate configuration through the latent space
Multi-fish Neuronal Model
Translating Activity from one Fish to Another
Multi-fish Neuronal Model
Translated Activity is Probable under the Recipient Model
Multi-fish Neuronal Model
Summary
- statistical model of whole-brain spontaneous neuronal activity → RBMs
- RBMs representation = composition of cell assemblies
- align the space of representation across multiple fish
- translate activity from one brain to another
only static
⠀
description
What about the dynamics?
Dynamics of Spontaneous Activity
Shared space of Representation
⠀
Dynamics of Spontaneous Activity
Segmenting the hidden space into a vocabulary of brain states
Dynamics of Spontaneous Activity
State sequence decoding from Neuronal Data
Dynamics of Spontaneous Activity
States Capture Large Scale Feature of Brain Activity
Dynamics of Spontaneous Activity
Across Individuals ?
- Are states stereoypical ?
- Are states used similarly ?
- Are states sequences stereotypical ?
Dynamics of Spontaneous Activity
States represent Stereotyped Brain States
Dynamics of Spontaneous Activity
States are Used Similarly by All Fish
Dynamics of Spontaneous Activity
Markovian Transition Rates are partially Conserved across fish
Dynamics of Spontaneous Activity
Markovian Dynamics - sub-structure and hub-states
Dynamics of Spontaneous Activity
Markovian Dynamics - sub-structure and hub-states
Conclusion & Perspectives
Goal : Is Spontaneous Brain Activity stereotypical across individuals?
- Spontaneous Activity is challenging to compare between individuals
- RBMs provide a representation of whole brain activity as a composition of cell assemblies
- Method : align this representation across multiple zebrafish larvae
- Method : define shared brain states and compare their temporal organization
- Spontaneous Brain Dynamics is partially conserved across individuals
- Joint training of RBMs (instead of Teacher/Student)
- Integrate a temporal model directly into the RBM (eg. HMM)
- Joint neuronal and behavioral studies to improve model interpretation
Shared Behavioral and Neuronal Representation
Zebrafish Reorientation
Ethics
SciComm
Thanks
Supplementary
Section Title
Smaller subtitle 1
$$\mathcal{U}(x) = \frac{1}{2}\gamma_+ x_+^2 + \frac{1}{2}\gamma_- x_-^2 + \theta_+x_+ + \theta_-x_-$$Section Title
Smaller subtitle 2
hey hey heeySection Title
Smaller subtitle 3
Anything goes here: text, images, charts…